

染色質組裝機制
2021/08- Assistant Professor, Institute of Biochemistry, National Chung Hsing University, Taiwan
2017/10–2019/09 博士後研究員 英國萊斯特大學
2015/05–2017/09 博士後研究員 英國帝國理工大學
2012/11–2015/04 博士後研究員 英國癌症研究中心
免疫缺陷、著絲粒不穩定、面部異常(immunodeficiency, centromeric instability, facial anomalies, ICF)綜合症是一種罕見的體染色體隱性遺傳疾病[1],主要特徵就如同名稱一樣為不同程度的免疫缺陷、著絲粒不穩定以及面部發育異常,由於免疫缺陷造成血清中缺少抗體,所以反覆感染為此病顯著的臨床表現[2, 3]。詳細的致病機制並不清楚,但是一般認為與DNA甲基化轉移酶3B(DNA methyltransferase 3B, DNMT3B)發生突變所造成的DNA甲基化減少(hypomethylation)有關[4, 5],截至目前的研究已證實DNMT3B、ZBTB24、CDCA7、HELLS四種基因與ICF綜合症發病有關[6]。根據致病基因的不同,ICF綜合症可細分為四種亞型,分別為ICF1(DNMT3B突變)、ICF2(ZBTB24突變)[7]、ICF3(CDCA7突變)以及ICF4(HELLS突變)[8],其中DNMT3B是最見的致病基因,大約有百分之五十的ICF患者帶有突變的DNMT3B基因。
截至目前的研究發現,zinc-finger and BTB domain containing 24(ZBTB24)是一個轉錄因子,其功能為調控cell division cycle associated 7(CDCA7)的表現[9],而CDCA7為一個染色質結合因子,它會促使helicase, lymphoid-specific(HELLS)被吸引到染色質上,並且誘導其活性進行染色質重塑(chromatin remodeling)[10],經重塑後而暴露出來的DNA就可以接著被DNMT3B進行甲基化,任何一個基因的突變都會造成有缺陷的DNA甲基化,進而導致ICF綜合症的發生。下圖是總結多年的研究之後所得到的最新作用模式:
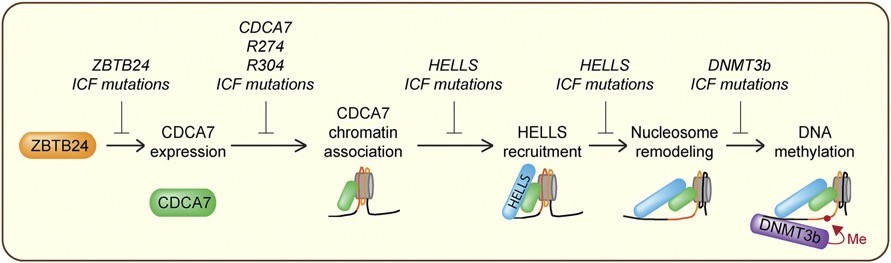
CDCA7是一個含有4CXXC zinc finger domain的蛋白質,此Zn-finger區域位於蛋白質的C端並且具有高度保留度,其主要功能為核酸結合。HELLS屬於SNF2 ATPase family中的SNF2-like subfamily,此次家族含有SMARCA2/4(BRM/BRG1)、CHD3-5(Mi-2)、CHD1/2、CHD6-9、SMARCA1/5(SNF2L/SNF2h,ISWI)以及ALC1等成員。大部分的成員不需要其他蛋白質的協助即具有染色質重塑活性,但是HELLS本身卻不具此活性,必須和CDCA7結合後形成CDCA7-HELLS ICF-related nucleosome remodeling complex(CHIRRC)才成為有活性的複合物(10)。CDCA7所扮演的角色為先利用其4CXXC zinc finger domain與核酸結合,再吸引HELLS到染色質並且刺激其染色質重塑活性,此功能對於後續的DNA甲基化攸關重要。
另一方面,有研究指出HELLS可以利用其N端的coiled-coil 區域直接與DNMT3B進行結合,再藉由DNMT3B來誘導DNA methyltransferase 1(DNMT1)與histone deacetylase 1(HDAC1)的結合[11]。此作用力對於DNMT3B的功能而言是非常重要的,因為DNMT3B必須作用在高度堆積的染色質區域,本身卻不具有染色質重塑的活性,因此勢必需要藉由HELLS的染色質重塑活性來消除此結構所造成的阻礙,才能夠順利進行DNA甲基化,因此了解HELLS與DNMT3B之間的詳細作用機制是非常重要的。
有鑑於此,未來的研究想利用生物化學實驗結合結構生物學方法來回答下列這些重要且未解的問題:
(1) CDCA7如何活化HELLS的染色質重塑活性?
(2) CHIRRC如何和染色質結合進行染色質重塑?
(3) HELLS與DNMT3B結合的詳細機制?
(4) ICF綜合症的突變研究?
參考文獻
1. P. Maraschio, O. Zuffardi, T. Dalla Fior, L. Tiepolo, Immunodeficiency, centromeric heterochromatin instability of chromosomes 1, 9, and 16, and facial anomalies: the ICF syndrome. J Med Genet 25, 173-180 (1988).
2. L. Tiepolo et al., Multibranched chromosomes 1, 9, and 16 in a patient with combined IgA and IgE deficiency. Hum Genet 51, 127-137 (1979).
3. M. M. Hagleitner et al., Clinical spectrum of immunodeficiency, centromeric instability and facial dysmorphism (ICF syndrome). J Med Genet 45, 93-99 (2008).
4. R. S. Hansen et al., The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A 96, 14412-14417 (1999).
5. G. L. Xu et al., Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187-191 (1999).
6. M. Vukic, L. Daxinger, DNA methylation in disease: Immunodeficiency, Centromeric instability, Facial anomalies syndrome. Essays Biochem 63, 773-783 (2019).
7. J. C. de Greef et al., Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am J Hum Genet 88, 796-804 (2011).
8. P. E. Thijssen et al., Mutations in CDCA7 and HELLS cause immunodeficiency-centromeric instability-facial anomalies syndrome. Nat Commun 6, 7870 (2015).
9. H. Wu et al., Converging disease genes in ICF syndrome: ZBTB24 controls expression of CDCA7 in mammals. Hum Mol Genet 25, 4041-4051 (2016).
10. C. Jenness et al., HELLS and CDCA7 comprise a bipartite nucleosome remodeling complex defective in ICF syndrome. Proc Natl Acad Sci U S A 115, E876-E885 (2018).
11. K. Myant, I. Stancheva, LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol 28, 215-226 (2008).
- Wang, Z. A.*, Millard, C. J.*, Lin, C. L.*, Gurnett, J. E., Wu, M., Lee, K., Fairall, L., Schwabe, J. W. and Cole, P. A. (2020, Jun). Diverse nucleosome Site- Selectivity among histone deacetylase complexes. eLife, 9, e57663.
- Willhoft, O.*, Ghoneim, M.*, Lin, C. L.*, Chua, E. Y. D., Wilkinson, M., Chaban, Y., Ayala, R., McCormack, E. A., Ocloo, L., Rueda, D. S. and Wigley, D. B. (2018, Oct). Structure and dynamics of the yeast SWR1-nucleosome complex. Science, 362(6411), eaat7716.
- Golzarroshan, B., Lin, C. L., Li, C. L., Yang, W. Z., Chu, L. Y., Agrawal, S. and Yuan, H. S. (2018, Sep). Crystal structure of dimeric human PNPase reveals why disease-linked mutants suffer from low RNA import and degradation activities. Nucleic Acids Research, 46(16), 8630-8640.
- Lin, C. L., Chaban, Y., Rees, D. M., McCormack, E. A., Ocloo, L. and Wigley, D. B. (2017, Jul). Functional characterization and architecture of recombinant yeast SWR1 histone exchange complex. Nucleic Acids Research, 45(12), 7249- 7260.
- Rees, D. M., Willhoft, O., Lin, C. L., Bythell-Douglas, R. and Wigley, D. B. (2017, Jul). Production and Assay of Recombinant Multisubunit Chromatin Remodeling Complexes. Methods in Enzymology, 592, 27-47.
- Sanders, K.*, Lin, C. L.*, Smith, A. J.*, Cronin, N., Fisher, G., Eftychidis, V., McGlynn, P., Savery, N. J., Wigley, D. B. and Dillingham, M. S. (2017, Apr). The structure and function of an RNA polymerase interaction domain in the PcrA/UvrD helicase. Nucleic Acids Research, 45(7), 3875-3887.
- Huen, J., Lin, C. L., Golzarroshan, B., Yi, W. L., Yang, W. Z. and Yuan, H. S. (2017, Mar). Structural Insights into a Unique Dimeric DEAD-Box Helicase CshA that Promotes RNA Decay. Structure, 25(3), 469-481.