

細胞膜作用蛋白功能研究與高解析度分子結構
國立中興大學生科院EMI雙語化計畫執行長 2022/8/01~
國立中興大學生物化學研究所 副教授 2021/08~
國立中興大學生物化學研究所 助理教授 2013/08 -2021/07
英國同步輻射中心Diamond Light Source 博士後研究員 2010/01-2013/07
英國倫敦帝國理工學院 博士後研究員 2007/01-2009/12
膜蛋白結構與功能實驗室 Membrane Protein Laboratory
本實驗室專門研究細胞膜相關的蛋白質結構與功能,包含受質轉運蛋白(transporters),通道蛋白(channels),孔洞形成型毒素蛋白(pore-forming toxins)。我們利用重組DNA的方式誘導重組基因之表現,並設法讓此疏水性蛋白分子穩定存在於水溶液中,以利後續蛋白質X-光晶體繞射及(protein X-ray crystallography)冷凍電子顯微鏡(cryo-EM)的實驗。目前我們已獲得高解析度蛋白質分子結構模型,並且正在進行了膜蛋白構型變化與多聚體組裝的分子動態分析,藉此我們可以推測蛋白在細胞膜上的功能以及其作用機制。
細菌鉀離子通道蛋白KtrAB的結構與功能
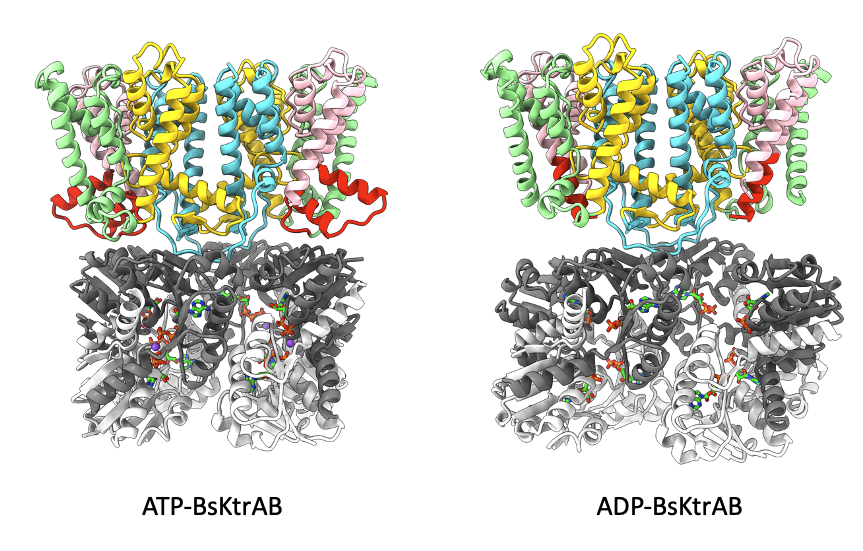
我們的研究團隊一直在研究整合膜蛋白的結構和功能,特別是K+通道。KtrAB是一個細菌K+通道,由K+孔KtrB和調節蛋白KtrA組成,KtrA屬於K+電導調節器(RCK)蛋白家族。KtrA以非共價的方式與KtrB結合。KtrA與各種細胞內信號分子的結合會觸發構象變化,然後傳播到通道孔,調節K+通道的門控。最近,我們證明ADP使KtrAB的K+流失活,但ATP激活該系統。令人驚訝的是,通過X光晶體繞射學和KtrA的熱穩定性研究顯示,在ATP依賴性激活機制中,Na+離子是至關重要的。我們解析了ATP和ADP-KtrAB覆合物的冷凍電子顯微鏡結構,從分子模型中揭示了近原子分辨率下的門控機制。此外,ATP和Na+對KtrAB系統的協同激活機制,可能可以幫助我們了解在中樞神經系統中,Na+激活的K+離子通道KNa的分子作用機制。關於KtrAB的研究成果已於2024年5月刊登於Nature Communications。https://www.nature.com/articles/s41467-024-48057-y
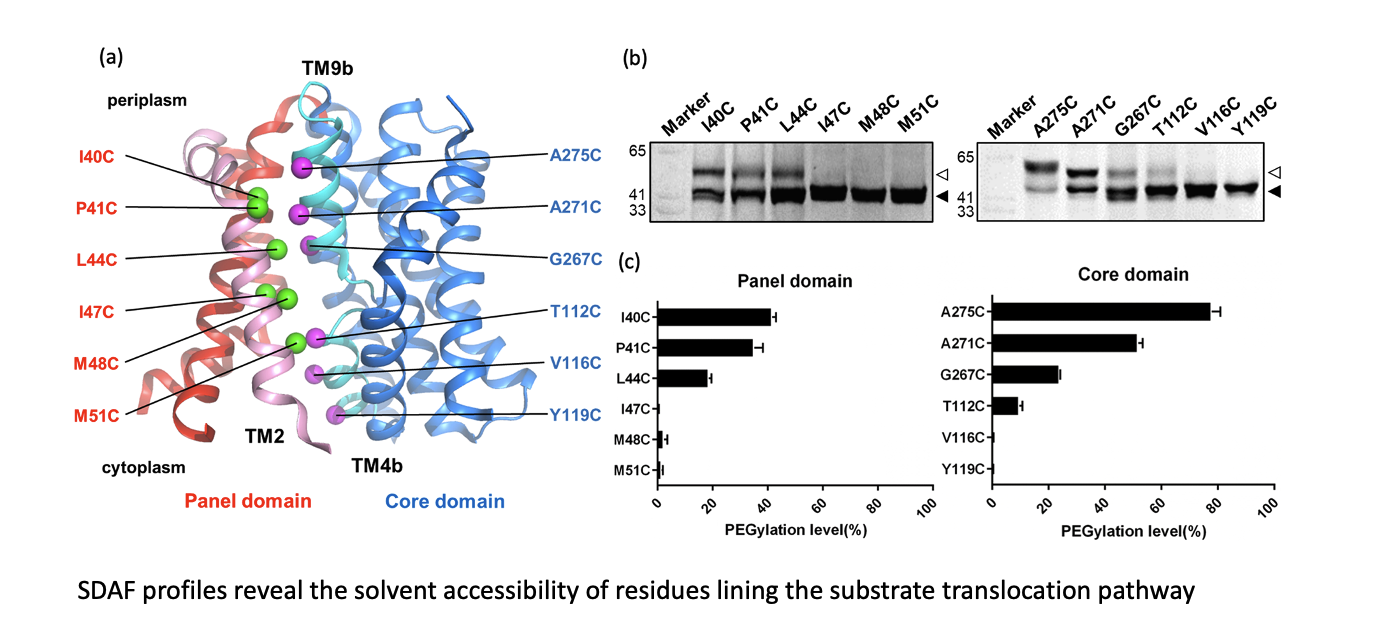
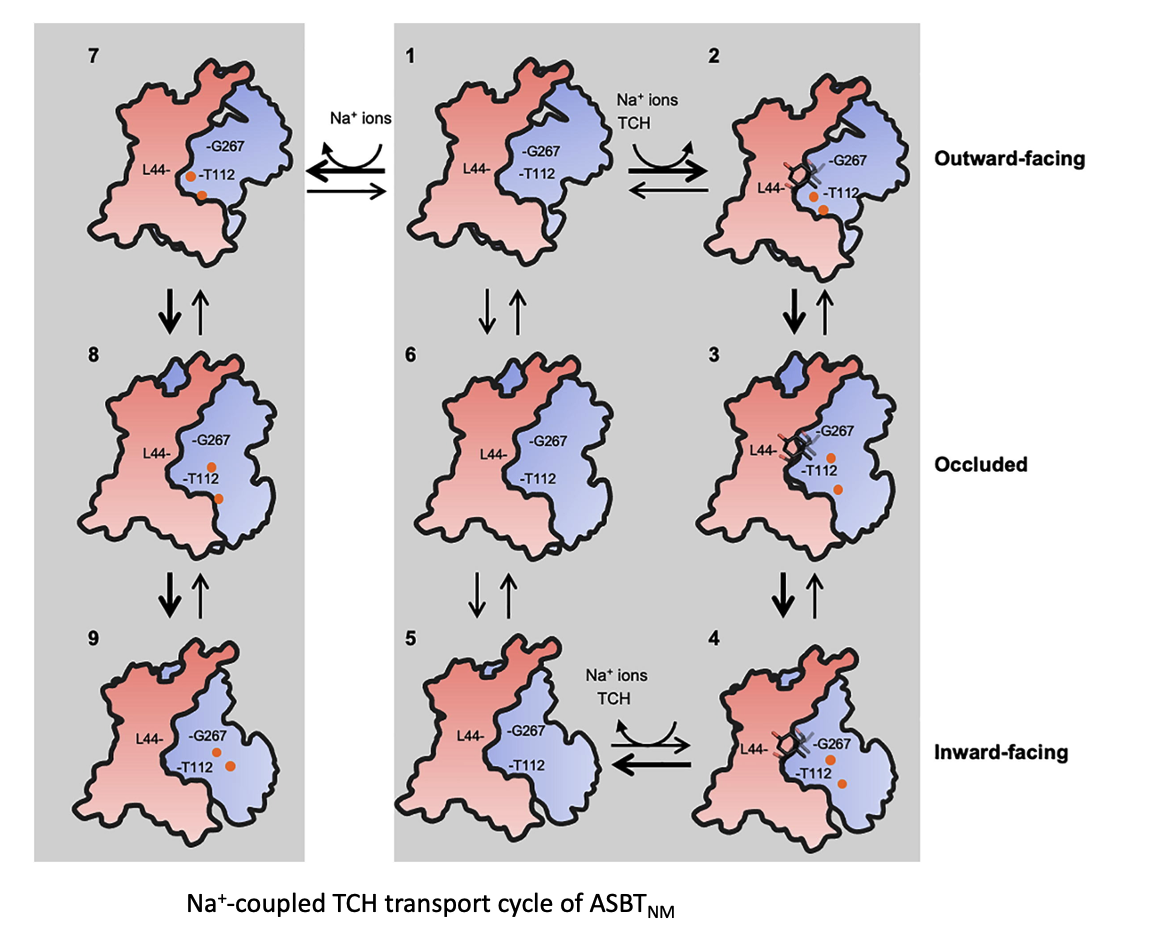
利用SDAF探針研究轉運蛋白構型改變
我們之前藉由GFP融合表現載體,將目標膜蛋白的C端融合GFP的技術,開發了一系列膜蛋白重組蛋白表現以及detergent穩定度的快速篩檢平台,稱為Fluorophore Absorption Size Exclusion Chromatography (FA-SEC),可以迅速優化膜蛋白表現條件,以及純化蛋白所使用的detergent,有利於後續蛋白子結構的研究。隨後我們又開發了Site-Directed Alkylation Detected by In-Gel Fluorescence (SDAF),有助於決定未知結構之膜蛋白的拓墣學(topology)結構。之後我們利用此技術應用在膜蛋白在細胞膜上構型改變之分子動態分析,以了解轉運受質的構型改變。
https://www.sciencedirect.com/science/article/pii/S0022283620306896?via%3Dihub#section-cited-by
https://www.nature.com/articles/s41598-019-49292-w
HuLab簡介 Prezi

英國愛丁堡大學結構暨分子生物學博士2004~2008
1. Jiang, D.-L., Yao, C.-L., Hu, N.-J., and Liu, Y.-C. (2021) Construction of a Tandem Repeat Peptide Sequence with Pepsin Cutting Sites to Produce Recombinant α-Melanocyte-Stimulating Hormone. Molecules 26
2. Hu, N. J., Li, S. Y., and Liu, Y. C. (2021) Recent Advances in Biocatalysis and Metabolic Engineering Catalysts 11, 1052
3. Lu, P. H., Li, C. C., Chiang, Y. W., Liu, J. H., Chiang, W. T., Chao, Y. H., Li, G. S., Weng, S. E., Lin, S. Y., and Hu, N. J. (2021) Dissecting the Conformational Dynamics of the Bile Acid Transporter Homologue ASBTNM. J Mol Biol 433, 166764
4. Huang, C. W., Lin, Y. S., Huang, W. C., Lai, C. C., Chien, H. J., Hu, N. J., and Chen, J. H. (2020) Inhibition of the clinical isolates of Acinetobacter baumannii by Pseudomonas aeruginosa: In vitro assessment of a case-based study. Journal of microbiology, immunology, and infection = Wei mian yu gan ran za zhi 1182
5. Wong, H. L., Hu, N. J., Juang, T. Y., and Liu, Y. C. (2020) Co-Immobilization of Xylanase and Scaffolding Protein onto an Immobilized Metal Ion Affinity Membrane. Catalysts 10, 1408
6. Fang, Y. T., Li, S. Y., Hu, N. J., Yang, J., Liu, J. H., and Liu, Y. C. (2020) Study on Cecropin B2 Production via Construct Bearing Intein Oligopeptide Cleavage Variants. Molecules 25
7. Lin, Y. H., Lin, S. Y., Li, G. S., Weng, S. E., Tzeng, S. L., Hsiao, Y. H., and Hu, N. J. (2019) Site-Directed Alkylation Detected by In-Gel Fluorescence (SDAF) to Determine the Topology Map and Probe the Solvent Accessibility of Membrane Proteins. Sci Rep 9, 13171
8. Liu, C. H., Chen, Y. T., Hou, M. H., Hu, N. J., Chen, C. S., and Shaw, J. F. (2018) Crystallographic analysis of the Staphylococcus epidermidis lipase involved in esterification in aqueous solution. Acta Crystallogr F Struct Biol Commun 74, 351-354
9. Tseng, W. H., Chang, C. K., Wu, P. C., Hu, N. J., Lee, G. H., Tzeng, C. C., Neidle, S., and Hou, M. H. (2017) Induced-Fit Recognition of CCG Trinucleotide Repeats by a Nickel-Chromomycin Complex Resulting in Large-Scale DNA Deformation. Angew Chem Int Ed Engl 56, 8761-8765
10. Huang, T. Y., Chang, C. K., Kao, Y. F., Chin, C. H., Ni, C. W., Hsu, H. Y., Hu, N. J., Hsieh, L. C., Chou, S. H., Lee, I. R., and Hou, M. H. (2017) Parity-dependent hairpin configurations of repetitive DNA sequence promote slippage associated with DNA expansion. Proc Natl Acad Sci U S A 114, 9535-9540
11. Lin, S. Y., Sun, X. H., Hsiao, Y.-H., Chang, S. E., Li, G. S., and Hu, N. J. (2016) Fluorophore Absorption Size Exclusion Chromatography (FA-SEC): An Alternative Method for High-Throughput Detergent Screening of Membrane Proteins. PloS one 11, e0157923
12. Hou, M. H., Chuang, C. Y., Ko, T. P., Hu, N. J., Chou, C. C., Shih, Y. P., Ho, C. L., and Wang, A. H. (2016) Crystal structure of vespid phospholipase A1 reveals insights into the mechanism for cause of membrane dysfunction. Insect Biochem Mol Biol 68, 79-88
13. Chia, J. Y., Tan, W. S., Ng, C. L., Hu, N. J., Foo, H. L., and Ho, K. L. (2016) A/T Run Geometry of B-form DNA Is Independent of Bound Methyl-CpG Binding Domain, Cytosine Methylation and Flanking Sequence. Sci Rep 6, 31210
14. Chang, C. K., Jeyachandran, S., Hu, N. J., Liu, C. L., Lin, S. Y., Wang, Y. S., Chang, Y. M., and Hou, M. H. (2016) Structure-based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol Biosyst 12, 59-66
15. Chin, K. H., Liang, J. M., Yang, J. G., Shih, M. S., Tu, Z. L., Wang, Y. C., Sun, X. H., Hu, N. J., Liang, Z. X., Dow, J. M., Ryan, R. P., and Chou, S. H. (2015) Structural Insights into the Distinct Binding Mode of Cyclic Di-AMP with SaCpaA_RCK. Biochemistry 54, 4936-4951
16. Hu, N. J., Iwata, S., Cameron, A. D., and Drew, D. (2011) Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 478, 408-411
17. Sonoda, Y., Newstead, S., Hu, N. J., Alguel, Y., Nji, E., Beis, K., Yashiro, S., Lee, C., Leung, J., Cameron, A. D., Byrne, B., Iwata, S., and Drew, D. (2011) Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure 19, 17-25
18. Weeratunga, S., Hu, N. J., Simon, A., and Hofmann, A. (2012) SDAR: a practical tool for graphical analysis of two-dimensional data. BMC Bioinformatics 13, 201
19. Weeratunga, S. K., Osman, A., Hu, N. J., Wang, C. K., Mason, L., Svard, S., Hope, G., Jones, M. K., and Hofmann, A. (2012) Alpha-1 giardin is an annexin with highly unusual calcium-regulated mechanisms. J Mol Biol 423, 169-181
20. Hu, N. J., Rada, H., Rahman, N., Nettleship, J., Bird, L., Iwata, S., Drew, D., Cameron, A., and Owen, R. (2014) GFP-based expression screening of membrane proteins in insect cells using the baculovirus system. Methods Mol Biol 1261, 197-209
21. Axford, D., Foadi, J., Hu, N. J., Choudhury, H. G., Iwata, S., Beis, K., Evans, G., and Alguel, Y. (2015) Structure determination of an integral membrane protein at room temperature from crystals in situ. Acta Crystallogr D Biol Crystallogr 71, 1228-1237
22. Hu, N. J., Yusof, A. M., Winter, A., Osman, A., Reeve, A. K., and Hofmann, A. (2008) The crystal structure of calcium-bound annexin Gh1 from Gossypium hirsutum and its implications for membrane binding mechanisms of plant annexins. J Biol Chem 283, 18314-18322
23. Hu, N. J., Bradshaw, J., Lauter, H., Buckingham, J., Solito, E., and Hofmann, A. (2008) Membrane-induced folding and structure of membrane-bound annexin A1 N-terminal peptides: implications for annexin-induced membrane aggregation. Biophys J 94, 1773-1781
24. Yusof, A. M., Hu, N. J., Wlodawer, A., and Hofmann, A. (2005) Structural evidence for variable oligomerization of the N-terminal domain of cyclase-associated protein (CAP). Proteins 58, 255-262
25. Hu, N. J., and Hofmann, A. (2005) Automated CD and fluorescence data processing with ACDP and AFDP (v2.4). Appl. Spectrosc. 59
26. Dabitz, N., Hu, N. J., Yusof, A. M., Tranter, N., Winter, A., Daley, M., Zschornig, O., Brisson, A., and Hofmann, A. (2005) Structural determinants for plant annexin-membrane interactions. Biochemistry 44, 16292-16300